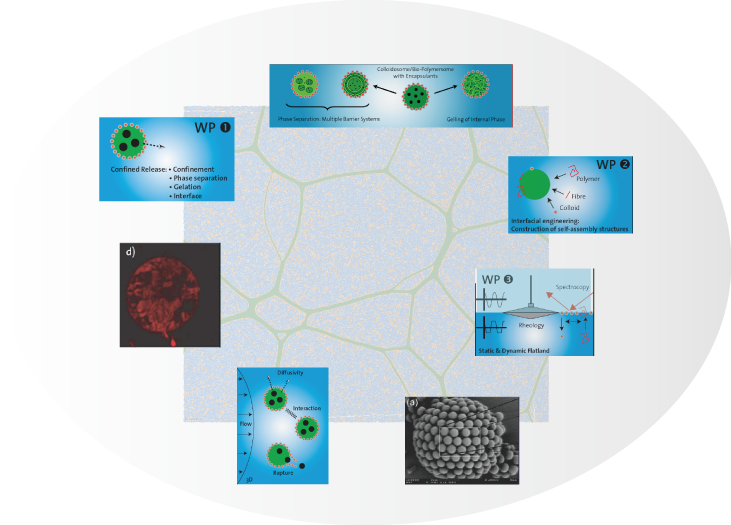
New controlled release systems produced by self-assembly of biopolymers and colloidal particles at fluid-fluid interfaces
| Mass transport in gels depends crucially on local properties of the gel network. We propose a method for identifying the three-dimensional (3D) gel microstructure from statistical information in transmission electron micrographs. The gel strand network is modelled as a random graph with nodes and edges (branches). The distribution of edge length, the number of edges at nodes and the angles between edges at a node are estimated from transmission electron micrographs by image analysis methods. The 3D network is simulated by Markov chain Monte Carlo, with a probability function based on the statistical information found from the micrographs. The micrographs are projections of stained gel strands in slices, and we derive a formula for estimating the thickness of the stained gel slice based on the total projected gel strand length and the number of times that gel strands enter or exit the slice. [hide]
Scientific Board
Scientific Stuff
Associated Scientists |
|
Enjoy your reading
SY Tee, AR Bausch, PA Janmey, | |
Selected conferences (co-)organized by project members8th World Congress on Computational Mechanics WCCM8 200830 June - 5 July 2007, Venice, Italy ► |
13 May 2025
![]() mk
mk